Research: Designed Sensors
Designed Phase-Changing COVID, Cancer and Bioterror Weapon Sensors
With funding from DARPA, the NIH and CDMRP, we are developing new design methods for extremely high signal-to-noise biosensing. These use designed supercharged phase-changing proteins that fold when they bind to small molecule or protein targets. Projects are ongoing aimed at detecting cancer biomarkers, biomarkers for COVID and car-T syndrome disease progression, chemical weapons, and proteins and small molecules involved in hormonal regulation.
There are two significant advances that we have made as a part of these projects:
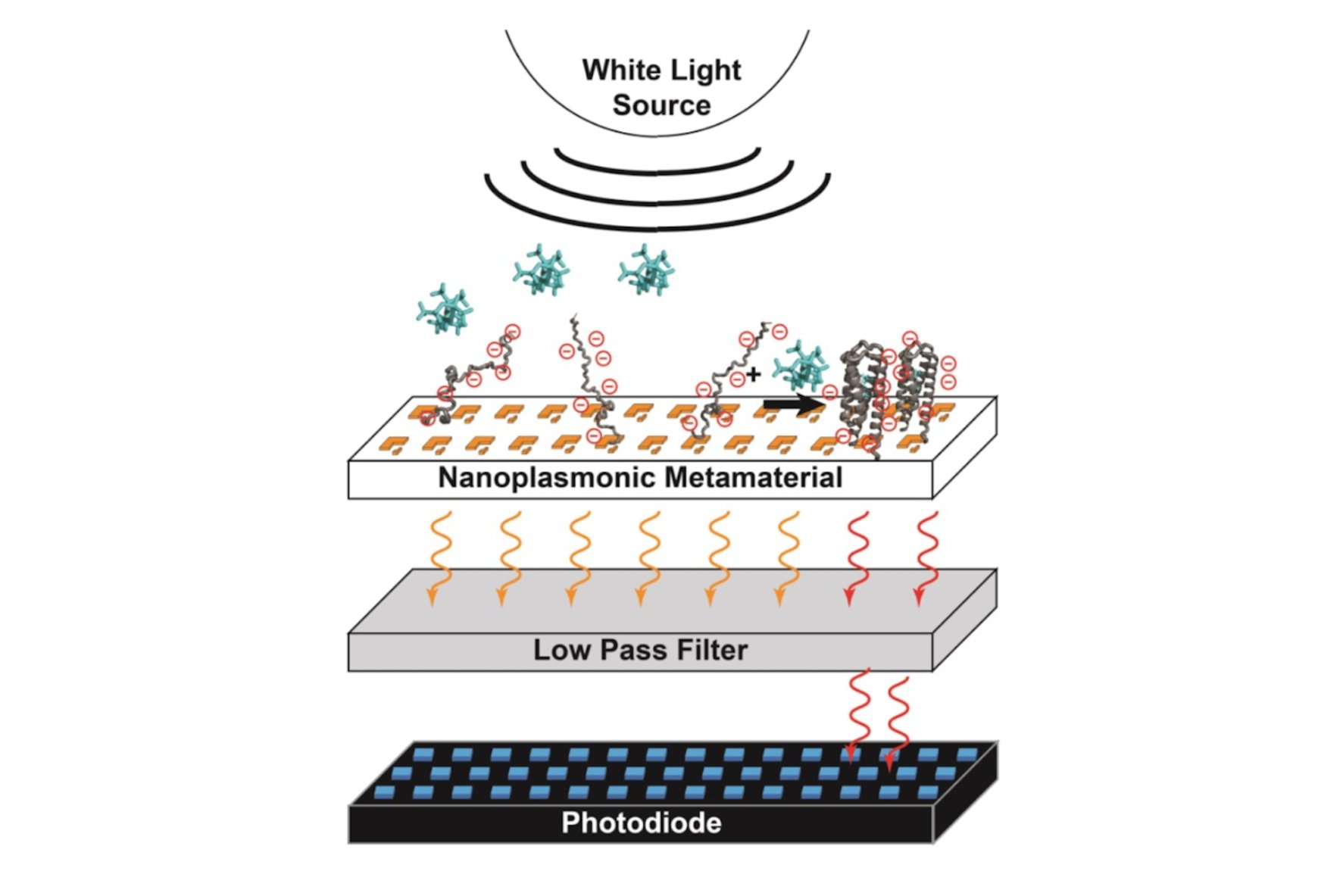
On-off sensing driven by ligand-induced folding of a supercharged sensor protein attached to a metamaterial transmission filter.
1. Protein surface supercharging of natural and designed proteins imparts a ligand induced folding mechanism that produces a greater than 1,000-fold increase in the change in the local electric fields upon ligand binding — as compared to the slight change in local dipoles caused by ligand binding to a folded protein, tens of full charges move from nanometers away to within angstroms from the sensing element. We have produced both plasmonic sensors and carbon nanotube-based sensors that are significantly higher S/N than the current state of the art for biosensing.
This project involves collaborations with Dan Heller from MSKCC for carbon nanotube-based detection and David Crouse and Alexander Khanikaev, electrical engineers at Clarkson University and CCNY respectively, for plasmonic metamaterial-based detection.
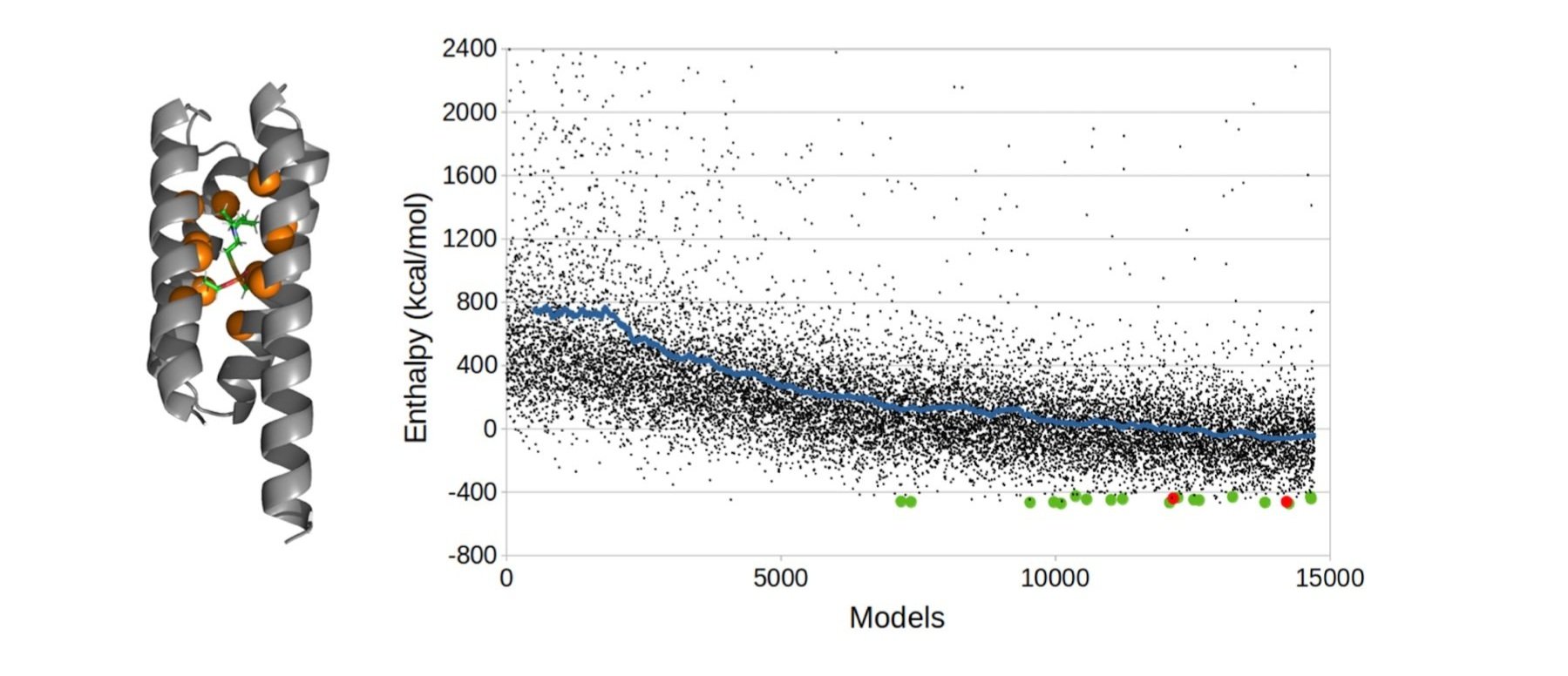
2. The large phase change enables the complete burial of small molecule binding sites within the core of the protein as opposed to binding ligands on the protein surface. The folded protein thus contacts the entire surface area of the protein, resulting in a higher interaction surface area, higher affinity, and greatly increased specificity. Our first paper on this, a selective, sensitive detector for VX nerve agent, was published in Science Advances.
This project involves machine learning algorithmic design in collaboration with Vikas Nanda at Rutgers University.
Computational design of a VX binding protein using a genetic algorithm.